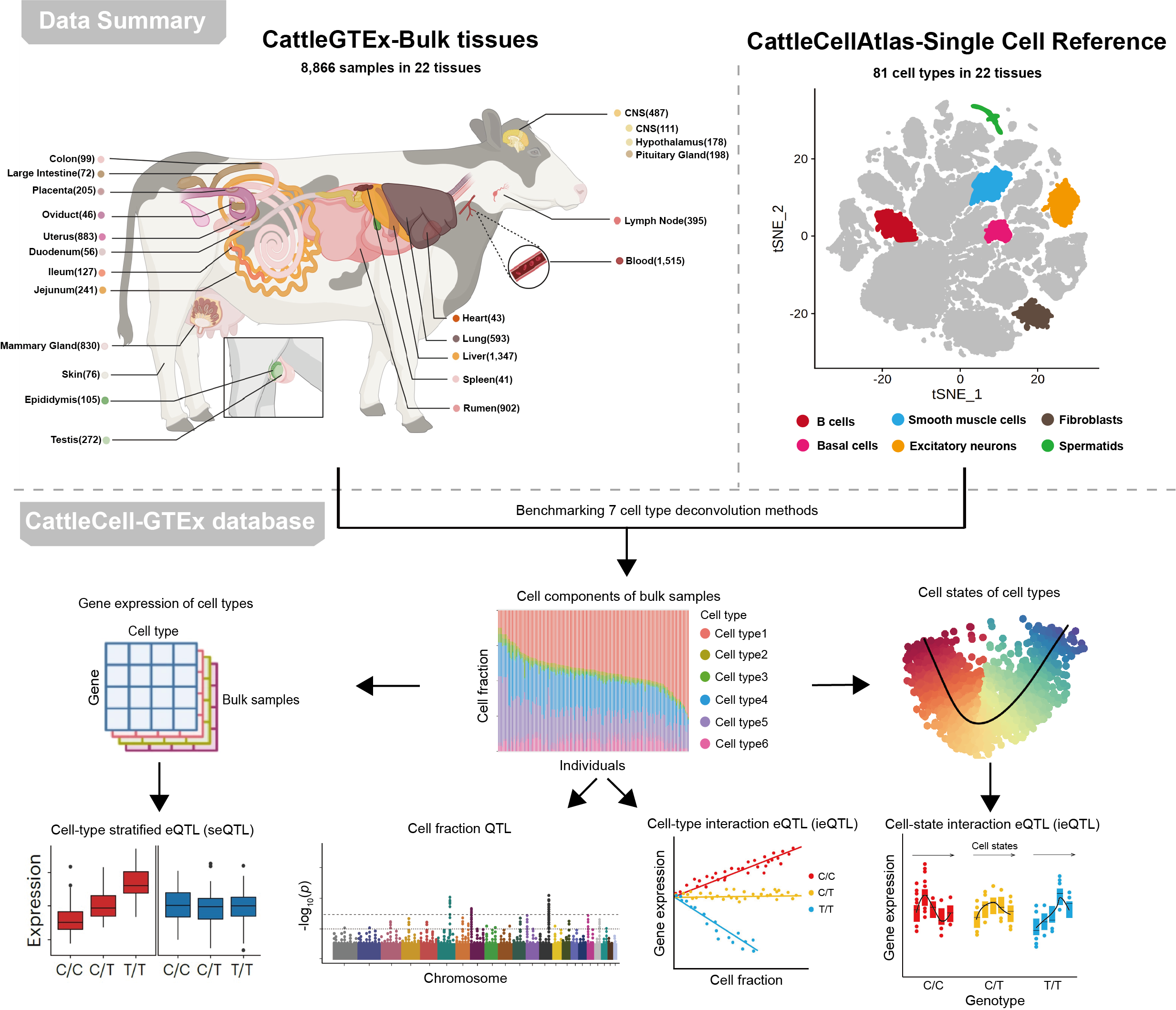
What is CattleCell-GTEx ?
Here, by integrating 8,866 bulk RNA-seq samples and 999,192 single cells of 72 cell types in 22 bovine tissues, we presented a comprehensive atlas of regulatory variants at the cell type resolution in cattle. This resource included 18,557 beGenes, 168 fQTL, 16,538 cseGenes, 16,341 cell-type ieGenes, and 15,012 cell-state ieGenes. It offers an in-depth understanding of the genetic regulatory effects on the bovine transcriptome at the cellular level and their impacts on complex traits and population selection in cattle. The CattleCell_GTEx will serve as an invaluable resource for cattle genomics, genetics, selective breeding, and domestication.
Please cite: Houcheng Li, Huicong Zhang, Pengju Zhao, Qi Zhang, Senlin Zhu, Tao Shi, Ya-Nan Wang, Jing-Sheng Lu, Liu Yang, John Francis O'Grady, David E MacHugh, Yu Wang, Zhenyu Wei, Xuemei Lu, Ming-Shan Wang, Bo Han, Weijie Zheng, Ao Chen, Shamima Akter, Nayan Bhowmik, Ying Ma, Ransom L. Baldwin VI, Congjun Li, Jicai Jiang, Li Ma, Christian Maltecca, Junjian Wang, Mian Gong, Xiaoning Zhu, Qing Lin, Yang Xi, Di Zhu, Jinyan Teng, Dailu Guan, Bingxing An, Jilong Ren, Yali Hou, Fei Wang, Bingjie Li, Laurent Frantz, Greger Larson, Zexi Cai, Goutam Sahana, Yu Jiang, Huizeng Sun, Dongxiao Sun, Geroge E. Liu, Lingzhao Fang. An atlas of cell type specific regulatory effects in cattle. BioRxiv, 2025. https://doi.org/10.1101/2025.06.23.661035